Get started
This guide provides step-by-step instructions for new users to get started with Tesorai Search. It covers the core workflow, including uploading mass spectrometry (RAW) and protein sequence (FASTA) files, launching search jobs with configurable parameters, and accessing job results.
Uploading files
Section titled “Uploading files”Uploading your data is the first step to using Tesorai Search. There are two main types of files you can upload:
-
RAW files: These are your mass spectrometry data files (e.g., Thermo
.raw). -
FASTA files: These contain protein sequence data for your search.
To upload files:
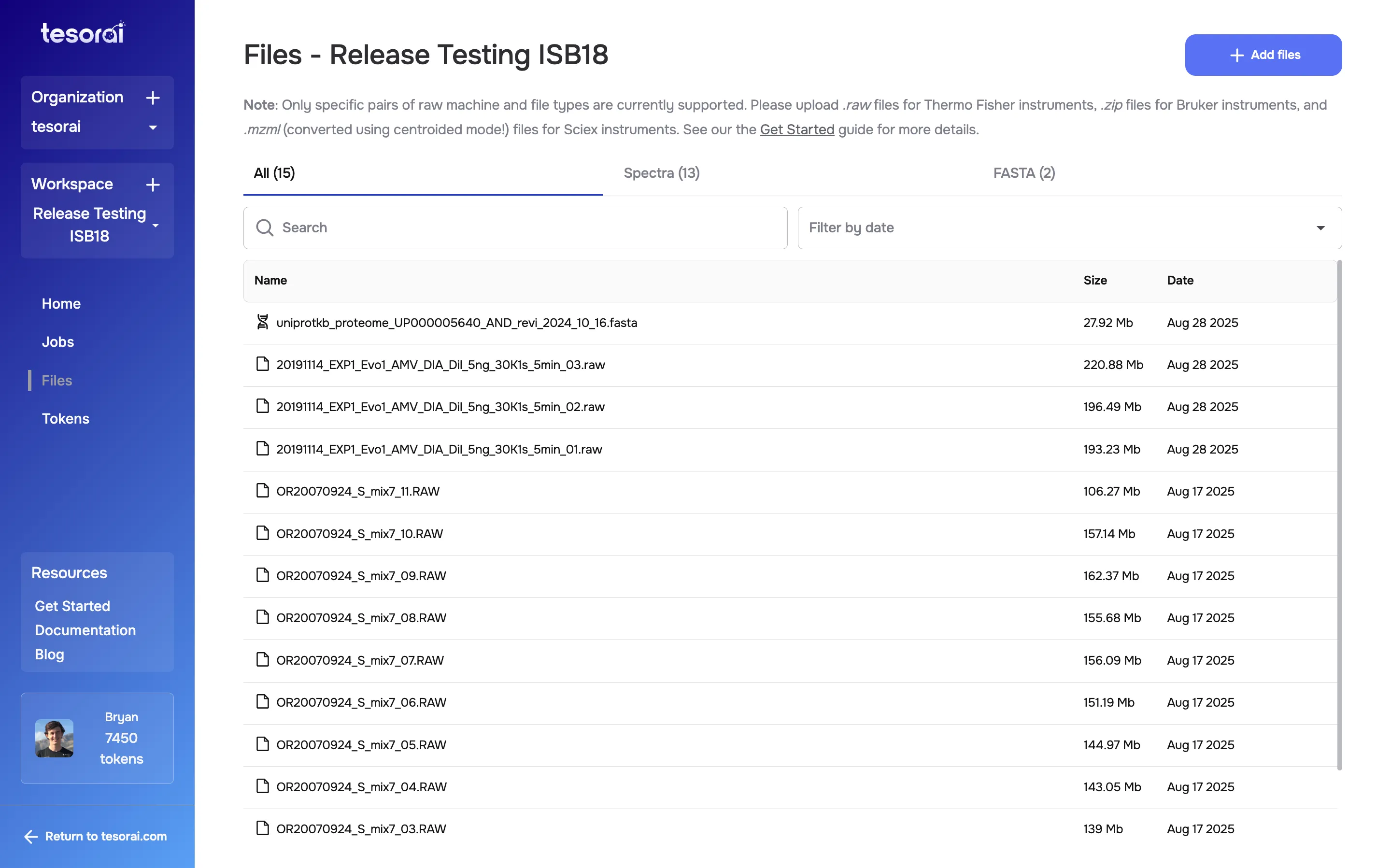
- Navigate to the Files page from the sidebar.
- Click the + Add files button in the top right or drag and drop
your files –
.raw,.mzml, or.zipfor spectra files, and.fastafor FASTA proteome files. - Uploaded files will appear in the list, organized by type:
SpectraorFASTA.
Running search jobs
Section titled “Running search jobs”Once your files are uploaded, you can launch a search job:
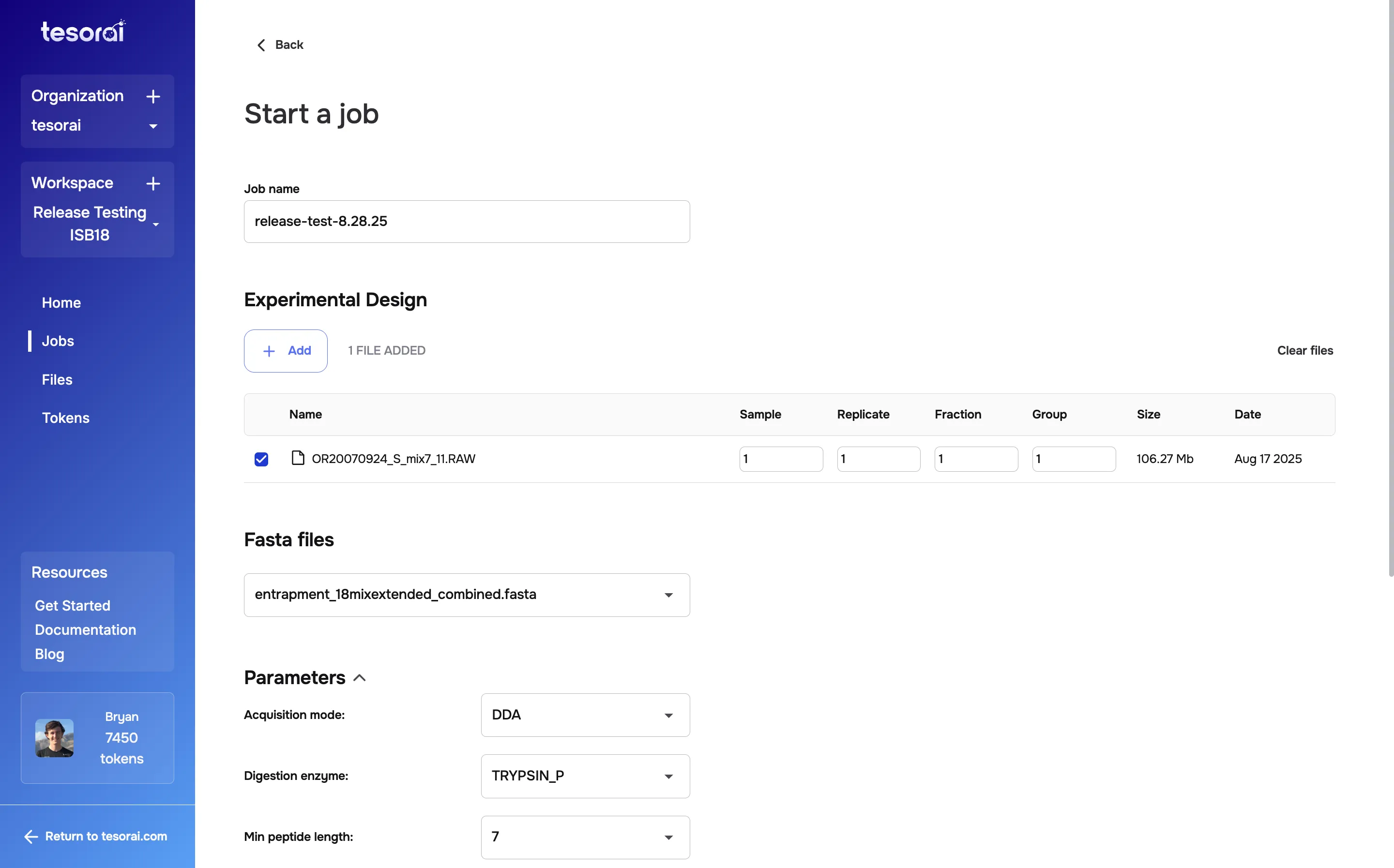
- Go to the Jobs page and click + Start job.
- Select a FASTA file and one or more RAW files.
- Configure your search parameters as needed (DDA/DIA, TMT-plex, variable modifications, FDR threshold, etc.).
- Click Start job to launch the analysis.
The job will appear in the Jobs list, where you can monitor for completion. You will also receive an email notification when the job is complete.
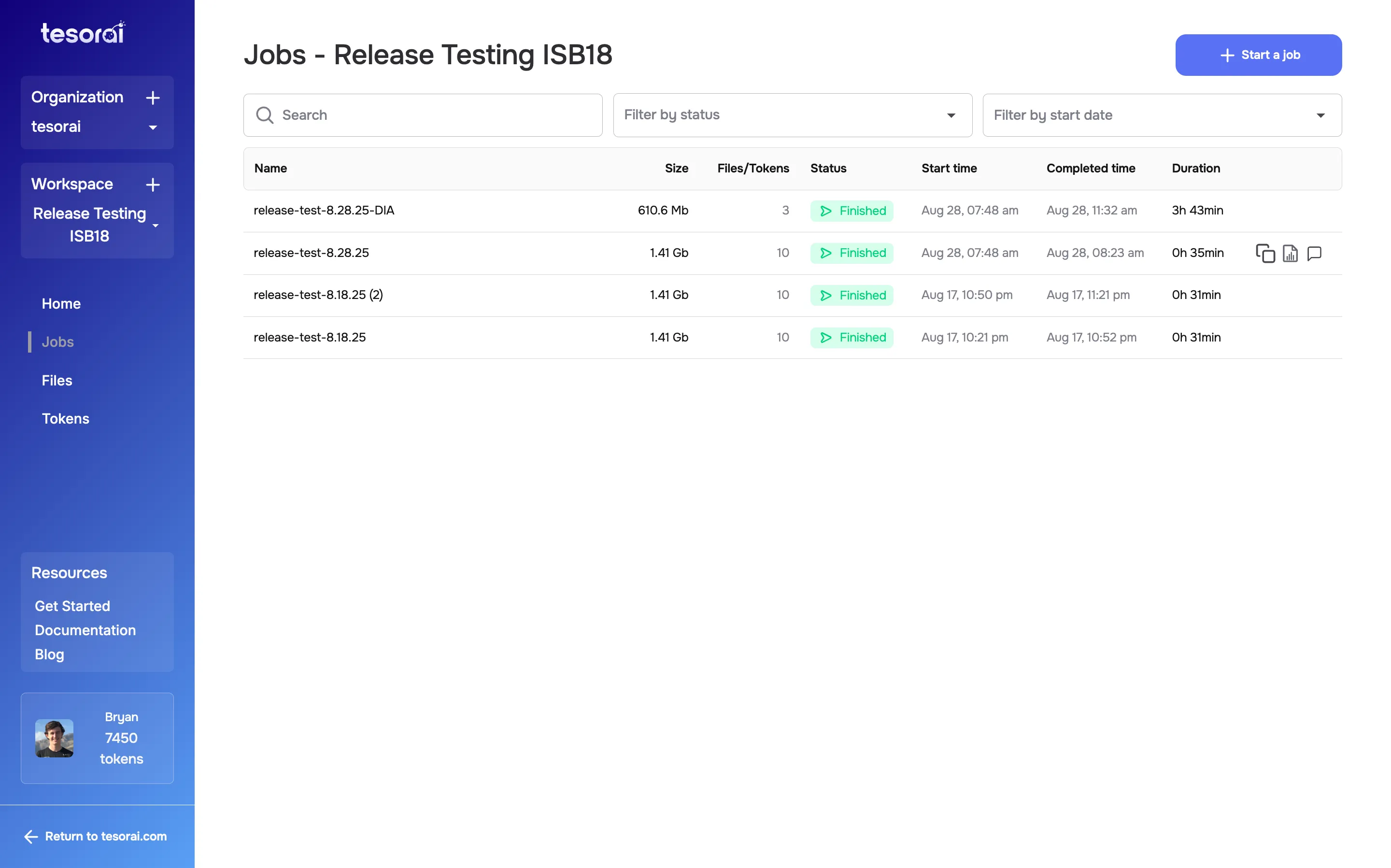
Viewing and Downloading Results
Section titled “Viewing and Downloading Results”Once the job is complete, you can view the results by clicking on the job or by clicking the ‘report’ icon on the right side of the job in the Jobs list.
![]()
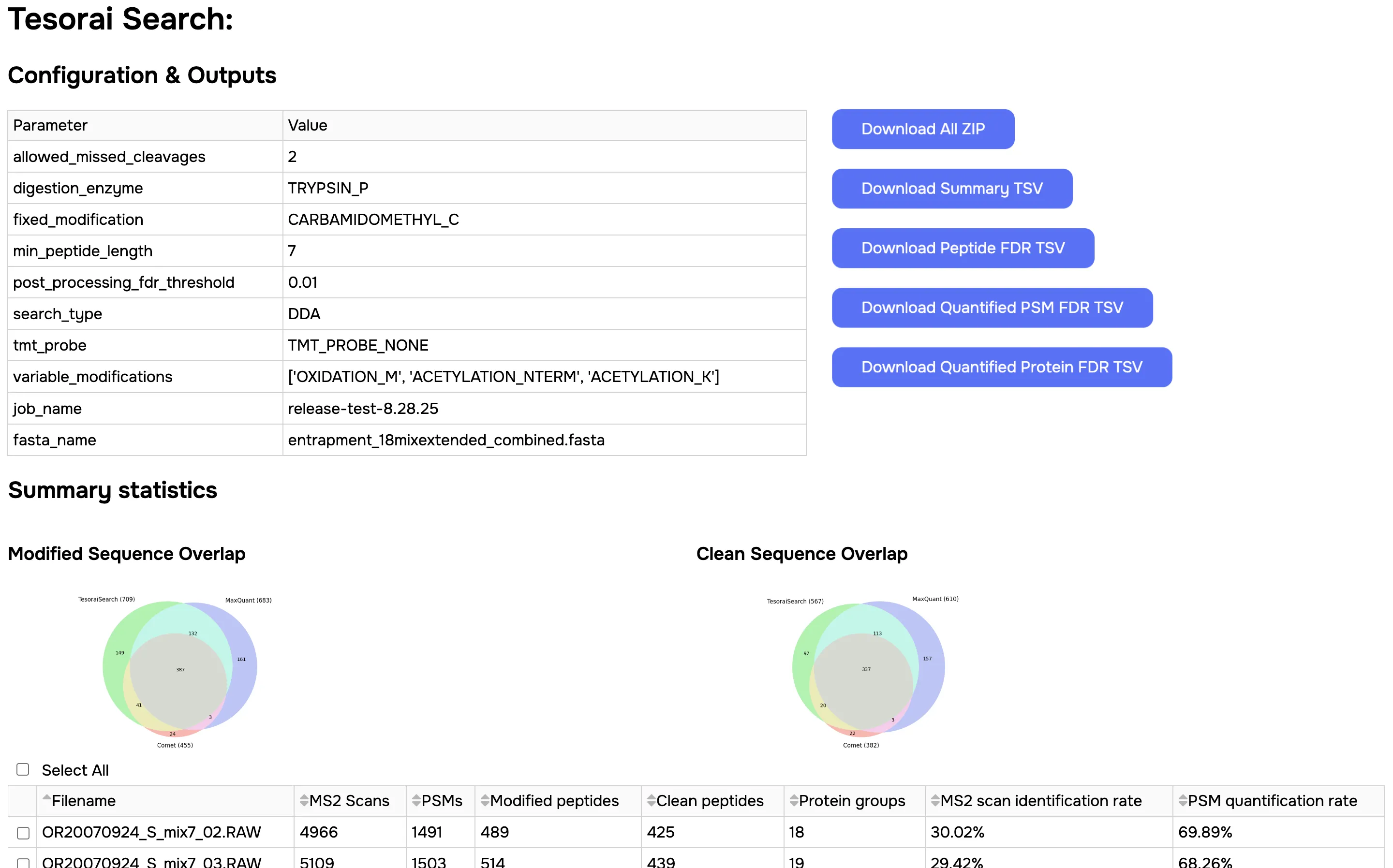
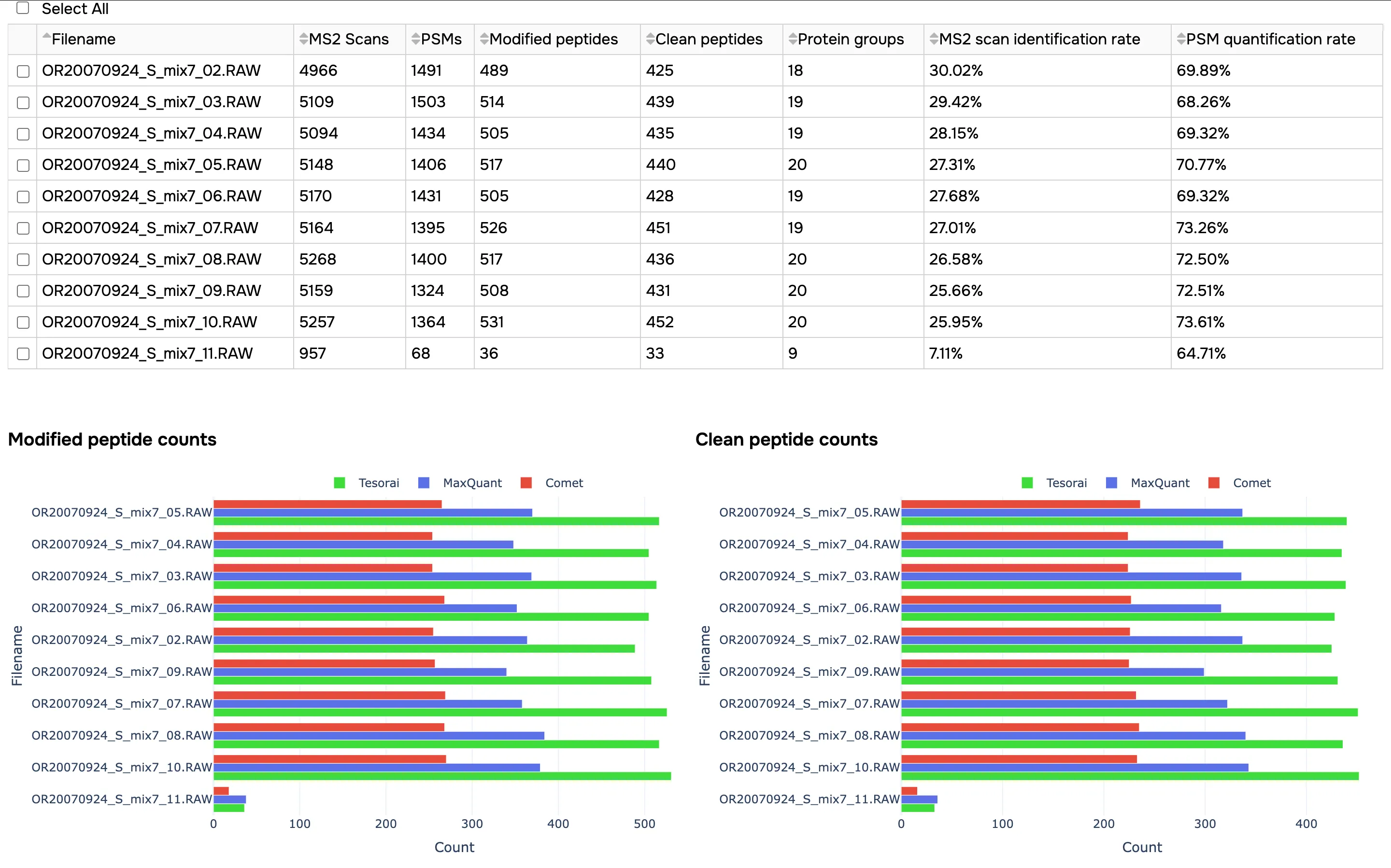
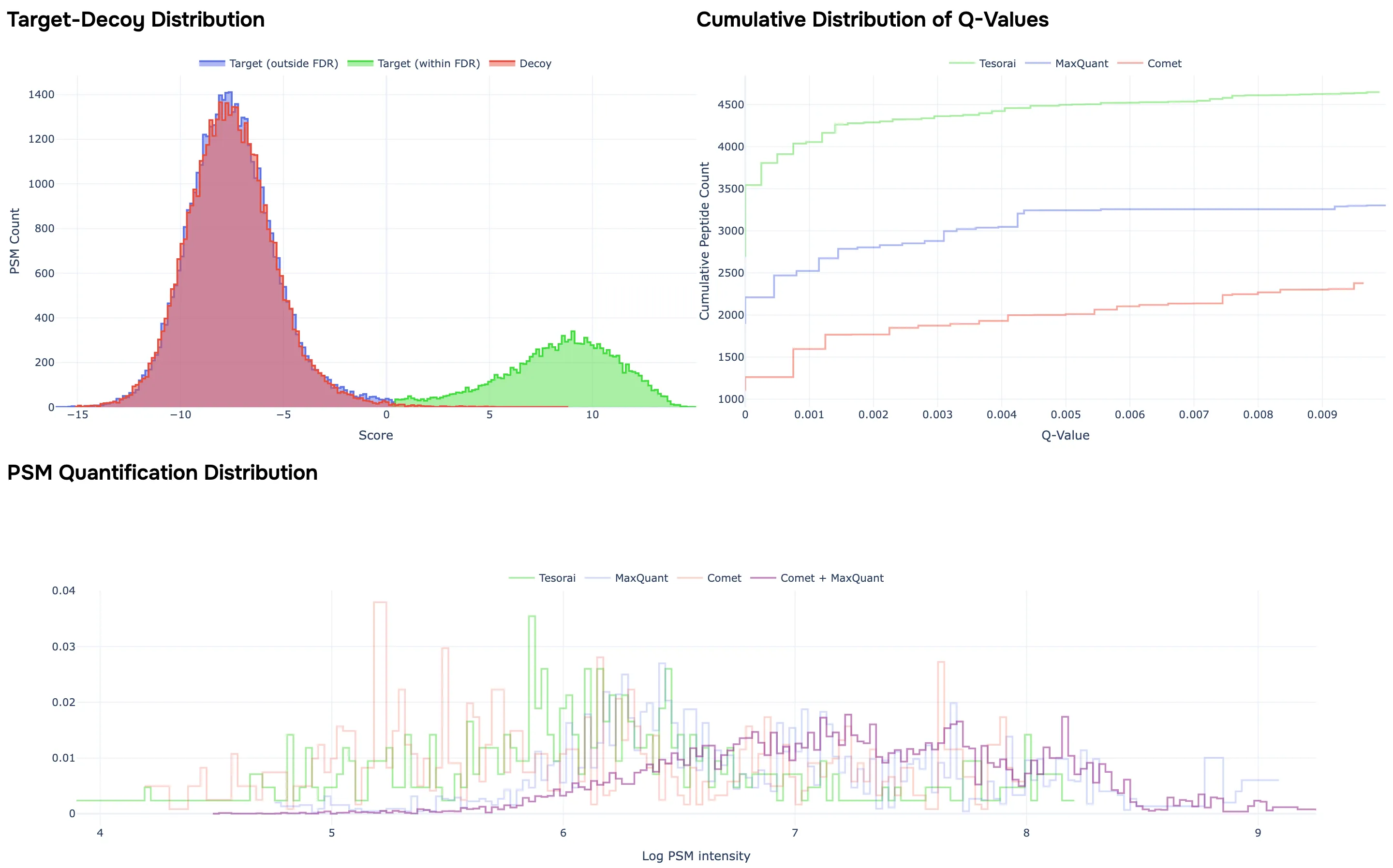
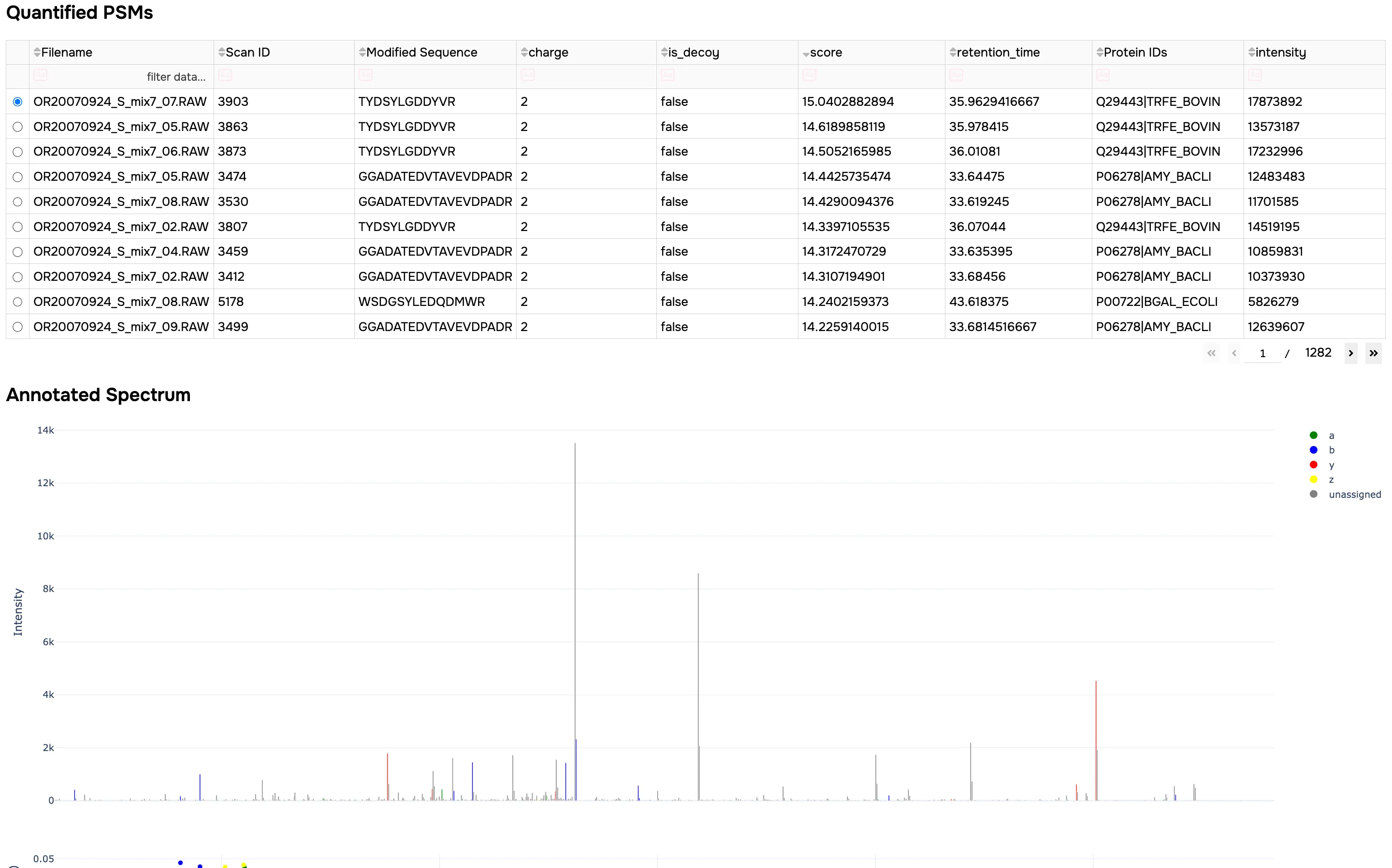
This will provide you with an overview report of the job, including:
- Summary statics from the search results
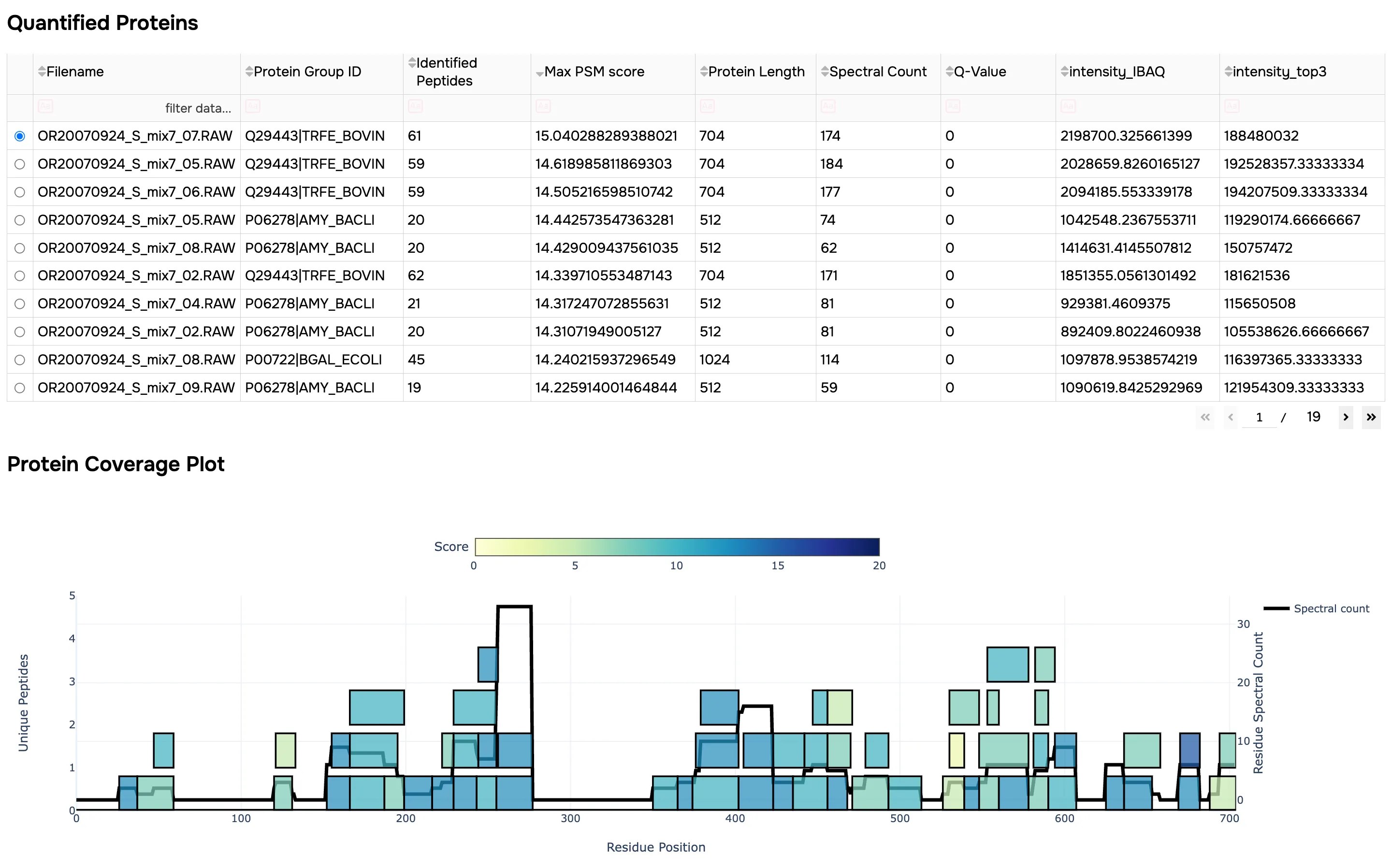
- Standard QC plots
- Searchable quantified PSM and protein tables, filtered to the specified FDR
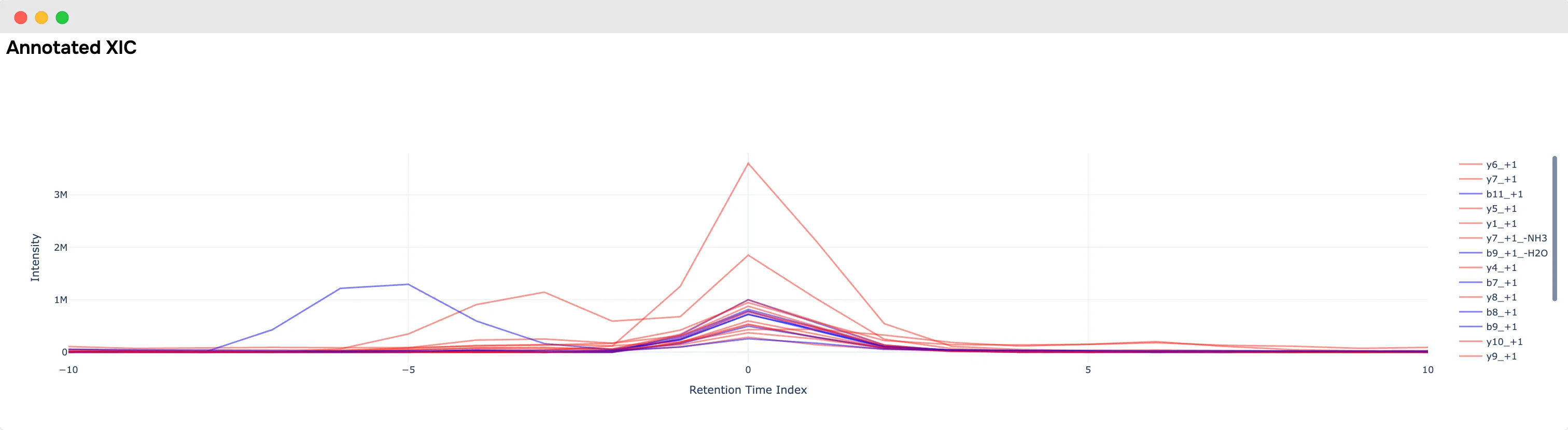
- Annotated spectrum viewer for PSMs and XIC viewer (DIA only)
- Protein coverage plots
The dashboard will also contain links (blue buttons in the upper-right corner) to download the Search results.
Note: You can search the Quantified PSMs and Quantified Proteins tables by:
- Entering a substring to match in the search bar at the top of any string-type
columns, e.g.
SGSEGSto match a peptide sub-sequence. - Entering
= <numeric_value>to match rows where the column value is equal to the specified numeric value. - Entering
> <numeric_value>to match rows where the column value is greater than the specified numeric value. - Entering
< <numeric_value>to match rows where the column value is less than the specified numeric value. - Entering
>= <numeric_value>to match rows where the column value is greater than or equal to the specified numeric value. - Entering
<= <numeric_value>to match rows where the column value is less than or equal to the specified numeric value.