Proteomics is one of the most data-rich fields in biology. Mass spectrometry experiments can capture thousands of proteins across samples, but transforming that raw output into biological insight usually requires a bioinformatician—and a fair bit of coding. At Tesorai, we’re changing that. With Tesorai Chat and Knowledge Graph, you can now go from spectra to pathways, no code required. When we think about proteomics data analysis, we think of it as a sequence of 3 steps that researchers typically need go through.
1. Primary Analysis – Transforming the Raw data into peptide/protein-level tables
Section titled “1. Primary Analysis – Transforming the Raw data into peptide/protein-level tables”This is the first step: turning instrument output into interpretable protein/peptide abundance tables. Our Tesorai Search tool has made it easy and intuitive to turn raw data into clean, usable inputs for downstream work.
2. Secondary Analysis – Identify genes/proteins relevant to the hypothesis at hand
Section titled “2. Secondary Analysis – Identify genes/proteins relevant to the hypothesis at hand”Once the protein abundance data is in hand, most users want to do basic exploration. That includes:
- Quality control checks
- Summary statistics and visualizations
This tier has already been popular among our users. Many have used Tesorai Chat to generate boxplots, heatmaps, and volcano plots, helping them understand where the strongest signals are in their data. We’re now adding the ability to perform differential analysis directly on our platform, without any coding required.
3. Tertiary Analysis – Put identified genes/proteins in biological context.
Section titled “3. Tertiary Analysis – Put identified genes/proteins in biological context.”This is where data truly becomes discovery: mapping proteomic changes onto biological context. Researchers want to know not just which proteins change, but also what those changes mean.
That means performing differential analysis, and cross-referencing findings against:
- Curated pathways
- Gene set enrichment (GSEA)
- Molecular functions and GO terms
- Protein–protein interactions
- Disease and drug associations
- Published literature and experimental data
This last tier is the most challenging—and the most exciting. While it’s still a work in progress, we’re taking the first step towards making it available today with Knowledge Graph in Tesorai Chat.
What’s New Today
Section titled “What’s New Today”With our latest update, biologists can go well beyond basic stats. Knowledge Graph now supports:
- Differential analysis between conditions
- Volcano plots to visualize significant changes
- Pathway and GSEA analysis for functional interpretation
- Disease and drug insights to link proteomics findings to therapeutic relevance
- Protein–protein interaction networks to uncover hidden relationships
To showcase some of these features, we looked at a Parkinson’s Disease dataset
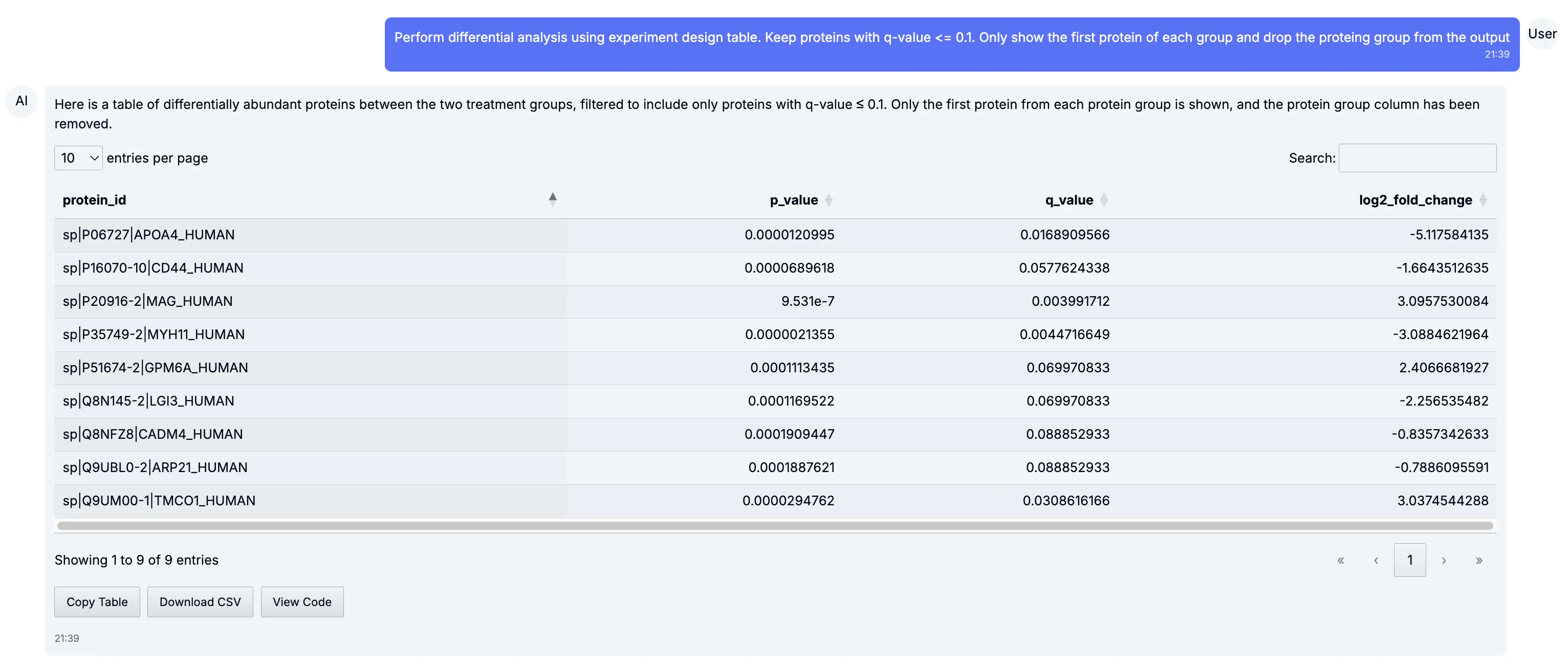
Differential analysis
Section titled “Differential analysis”To start, we ran a differential expression analysis between the Parkinson’s disease samples and controls. The volcano plot below highlights proteins that are both significantly different and strongly regulated, making it easy to spot potential biomarkers or candidates for follow-up.
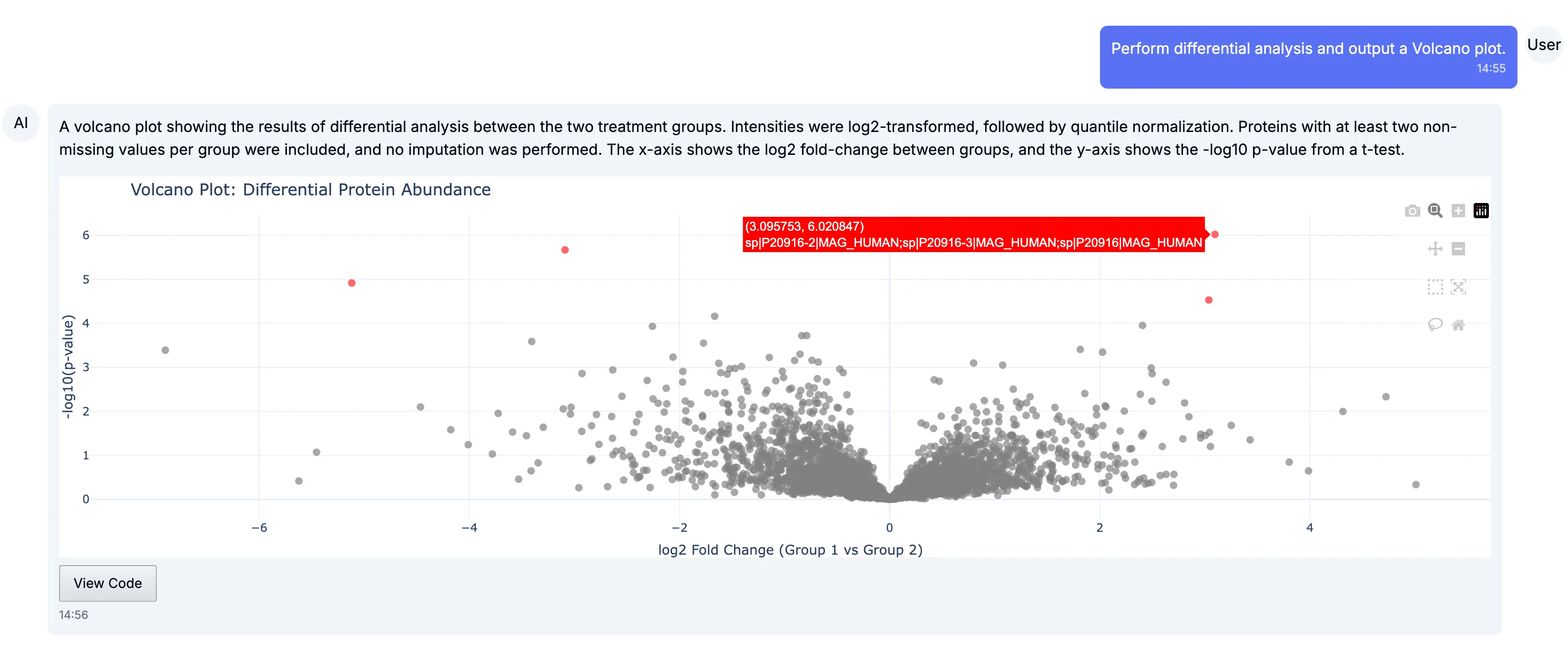
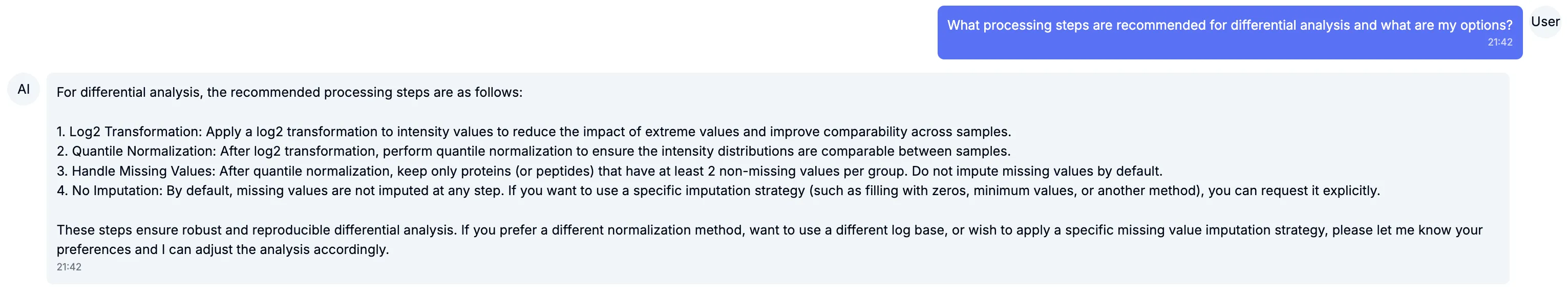
We’ve taken steps to ensure that differential analysis works well by default, without requiring specific knowledge of data imputation, data normalisation, statistical significance tests or multiple hypothesis corrections. For users who wish to provide specific instructions for any or all of these steps, we support different options they can decide what to use, or even ask Chat for benefits and drawbacks of each approach in their specific setting.
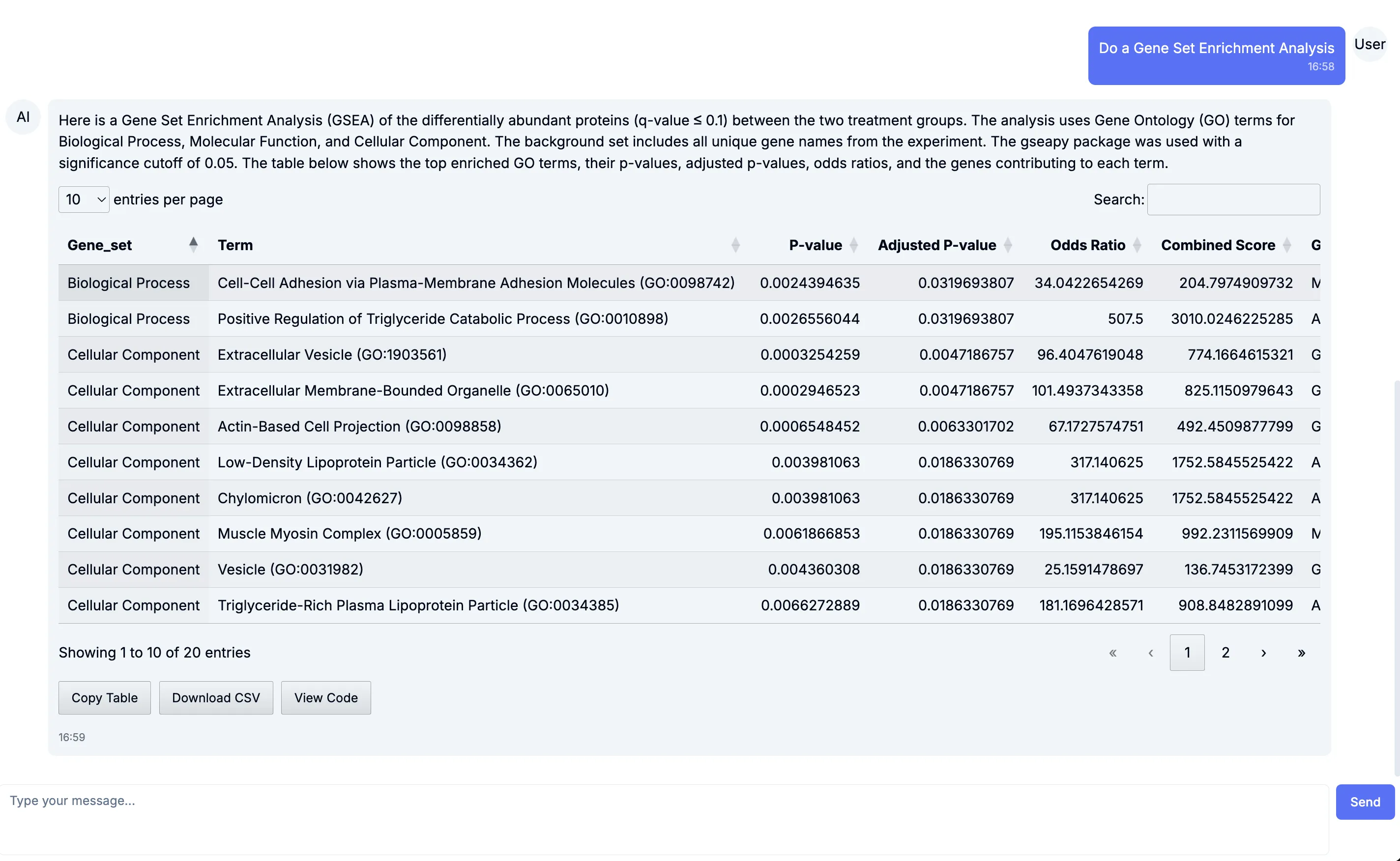
GSEA & Pathway analysis
Section titled “GSEA & Pathway analysis”Looking beyond individual proteins, we asked: Which pathways and processes are changing? Tesorai Chat runs GSEA and curated pathway analyses directly from your dataset. In this case, we can see enrichment of pathways relevant to neurodegeneration, providing biological context to the observed protein shifts.
The GSEA output further highlights groups of genes and proteins moving together, which is critical for understanding system-level effects.
Protein–protein interaction networks
Section titled “Protein–protein interaction networks”Of course, biology doesn’t happen in isolation. Proteins interact in complexes and pathways, and changes often ripple through networks. With a simple prompt, Tesorai Chat surfaces protein–protein interaction (PPI) networks built from curated databases.
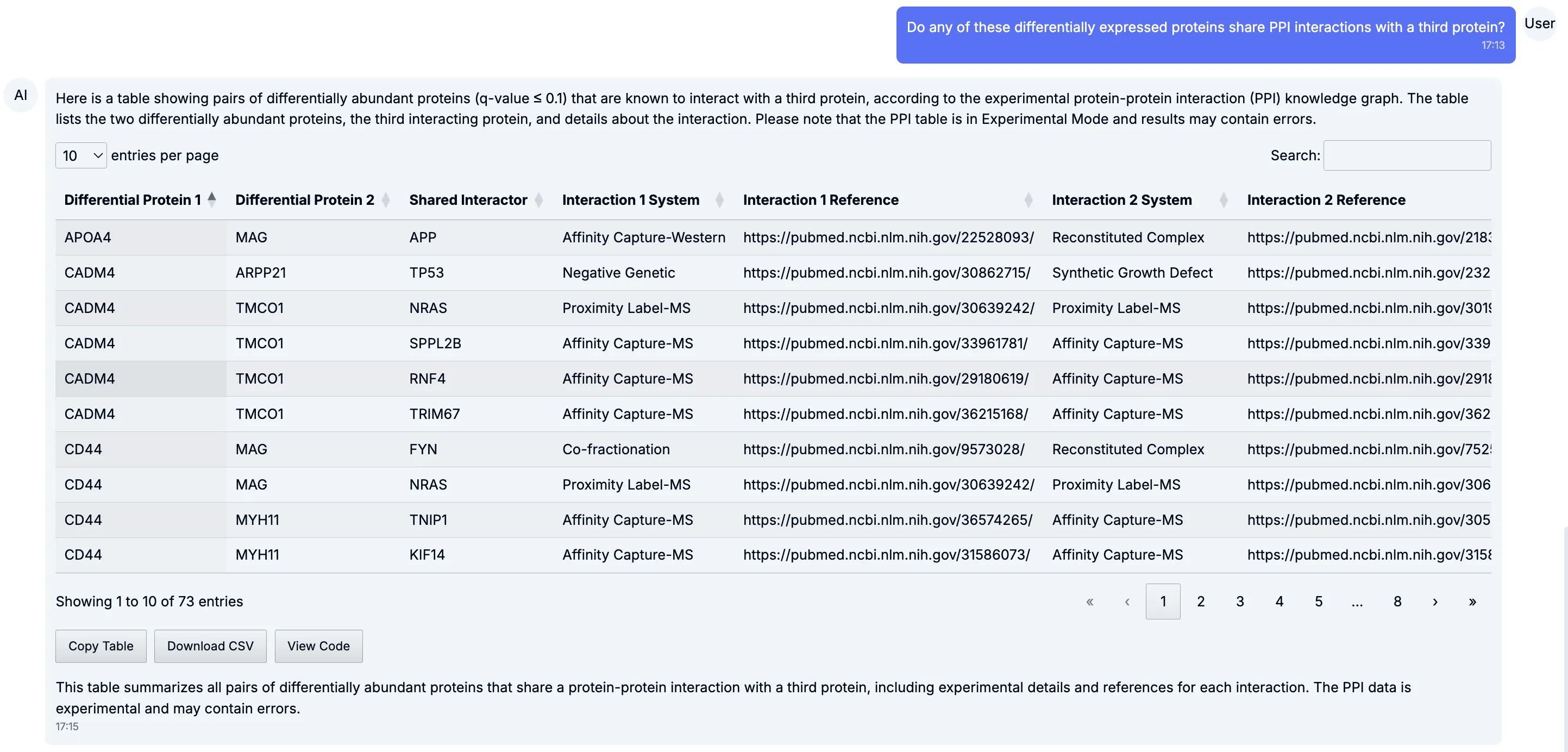
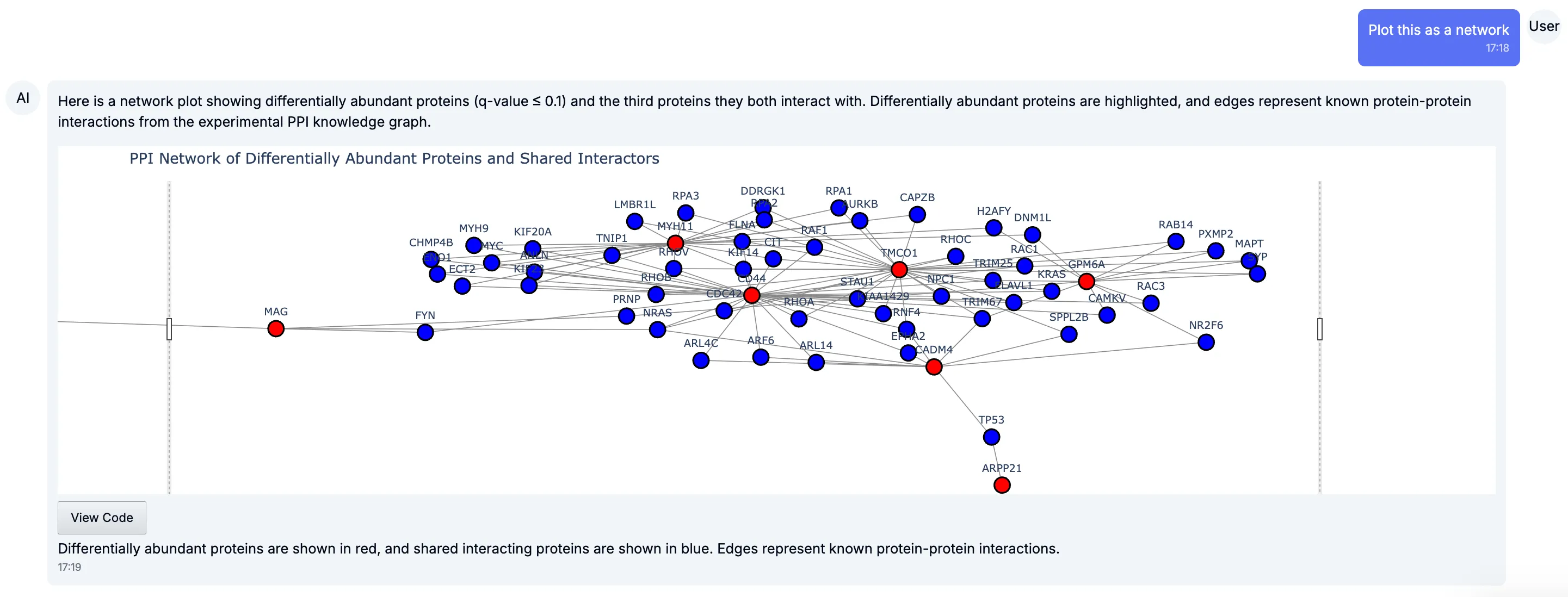
Zooming out, you can see how multiple regulated proteins connect into larger subnetworks — often pointing to functional hubs or modules worth investigating.
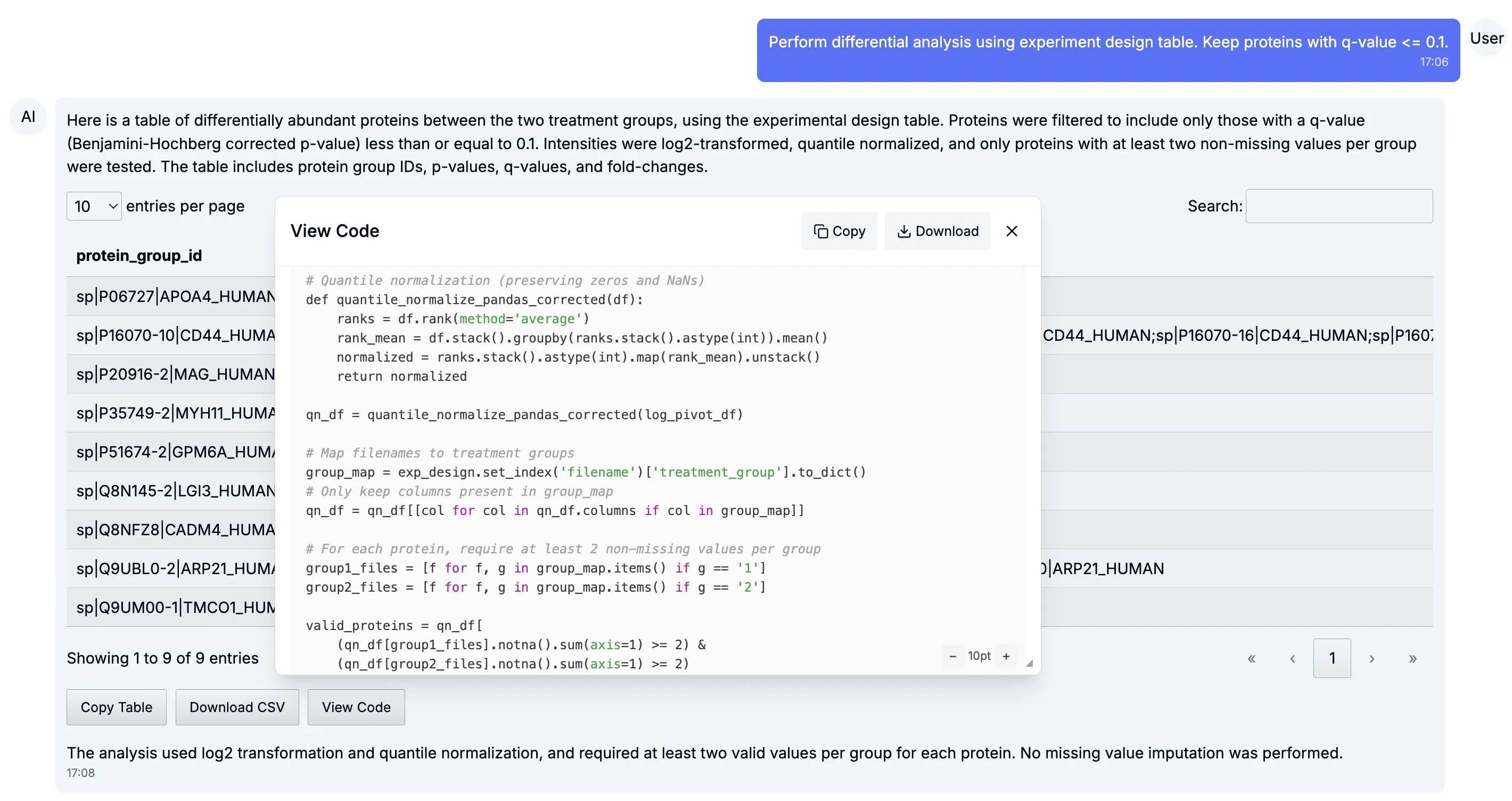
Code transparency
Section titled “Code transparency”Being able to audit and reproduce scientific analyses is critical. That’s why now, every figure and table generated by Tesorai Chat comes with transparent code.
This ensures reproducibility, lets you tweak analyses to your needs, and makes it straightforward to share methods in publications.
Why This Matters
Section titled “Why This Matters”By layering these three tiers of analysis into a conversational, no-code platform, we’re enabling biologists to move seamlessly from raw data to biological discovery.
- With Tesorai Search, you start by transforming raw spectra into usable tables.
- With Tesorai Chat, you explore and visualize your data quickly and interactively.
- With the Knowledge Graph, you connect your results to pathways, functions, and disease biology.
This integrated workflow brings proteomics closer to what it should be: a tool for discovery and insight, not a bottleneck of data processing.
Looking Ahead
Section titled “Looking Ahead”Tertiary analysis—cross-referencing proteomic results with the broader world of biology—is only just beginning in Tesorai Chat. As we expand our Knowledge Graph with more data modalities, scientific evidence and better knowledge curation, we’re building towards a future where your chat assistant becomes your virtual bioinformatician. We can’t wait to see what discoveries you’ll make.